The Bowhead whale genome resource
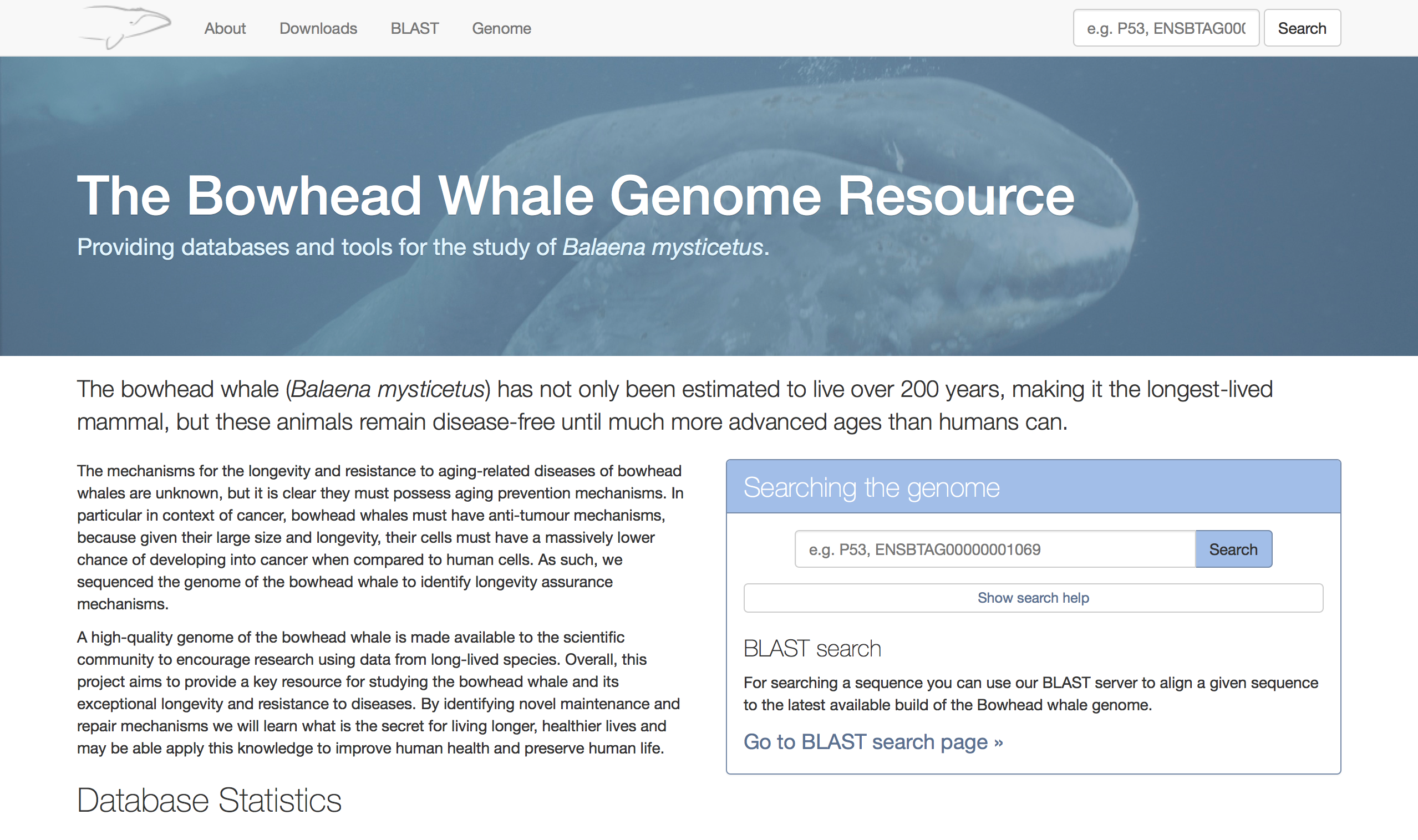
The Bowhead whale genome resource is a portal containing research conducted on the Bowhead whale. It is based heavily upon the Naked Mole-rat genome resource though it has had a number of changes to better suit the data.
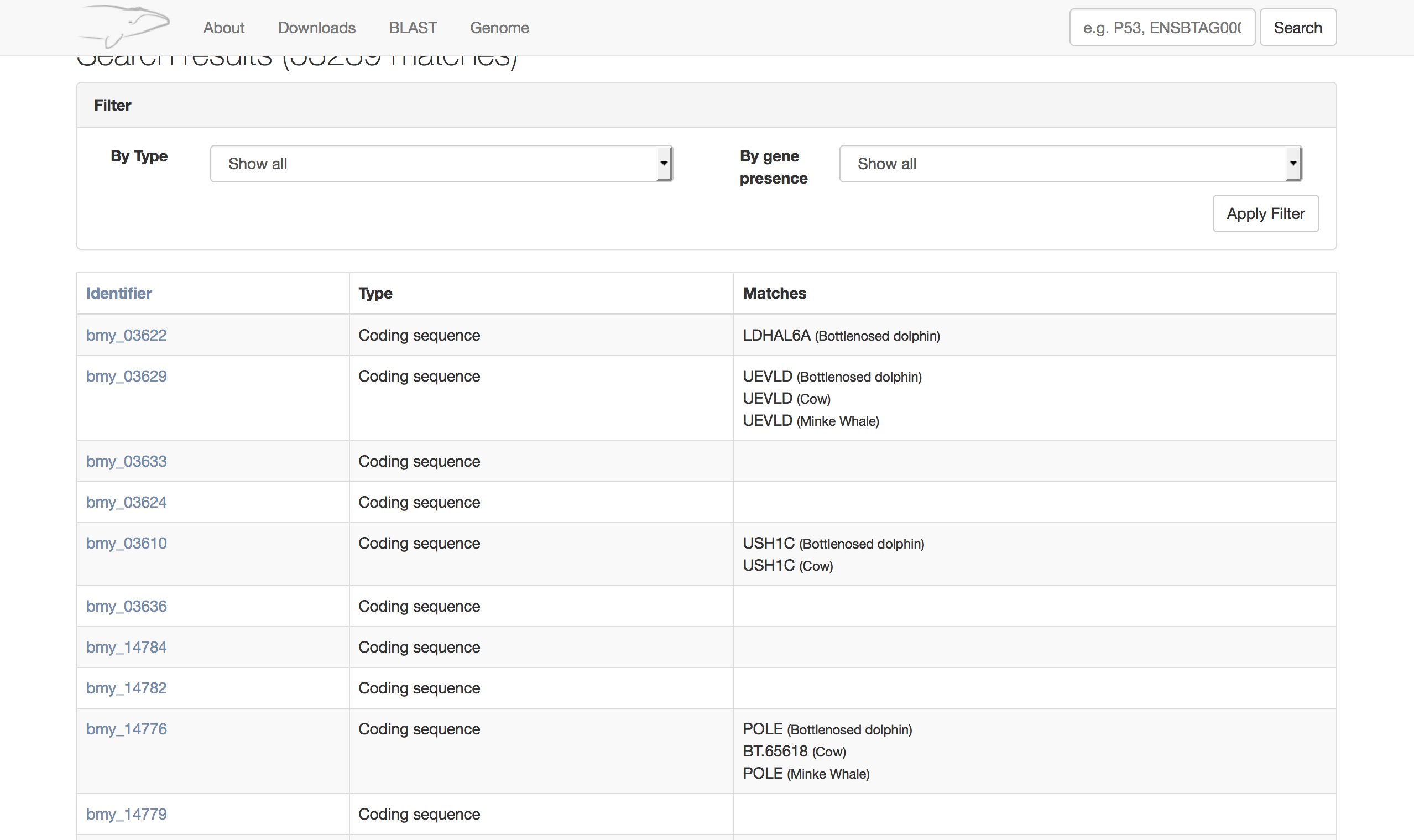
The data for the Bowhead whale is different so does not require best hits to be identified. Much like the Naked Mole-rat version it also uses the elasticsearch back end to to do the searching.
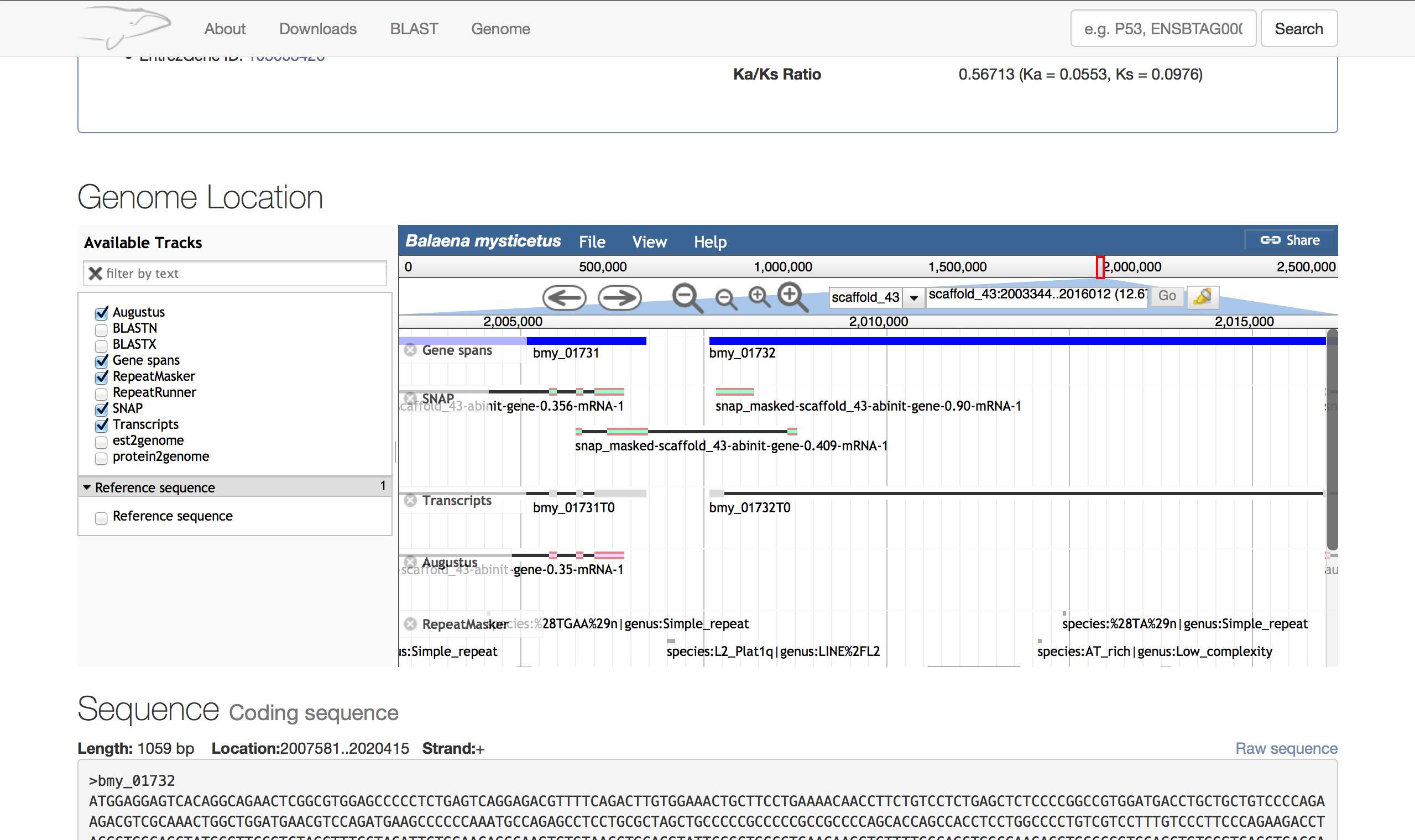
Processing a genome into the format the genome browser requires takes a significant amount of memory and with the Bowhead whale genome this proved to be difficult (it contains much more detail than the Naked Mole-rat). In this version the genome was processed using my own script and into a database; This database is then used to fetch the results for the genome browser. It is slightly slower than using flat files but later versions of the genome browser should eliminate this problem by caching data more effectively.